Machine Learning in Python using Scikit-Learn¶
The Dataset¶
- CAS Loss Reserve Database for Workers' Compensation, Medical Malpractice, and Private Passenger Auto
- Top 19 companies with remainder of industry as 20th observation
- Volume-weighted LDFs generated
import pandas as pd
ldf = pd.read_csv(
r'https://raw.githubusercontent.com/PirateGrunt/paw_rpm/master/notebooks/assets/links.csv',
index_col=['GRNAME','LOB'])
ldf.head()
What is Scikit-Learn?¶
Machine Learning in Python
- Simple and efficient tools for data mining and data analysis
- Accessible to everybody, and reusable in various contexts
- Built on NumPy, SciPy, and matplotlib
- Open source, commercially usable - BSD license
scikit-learn covers the majority of supervised and unsupervised ML techniques available today and is continually expanding¶
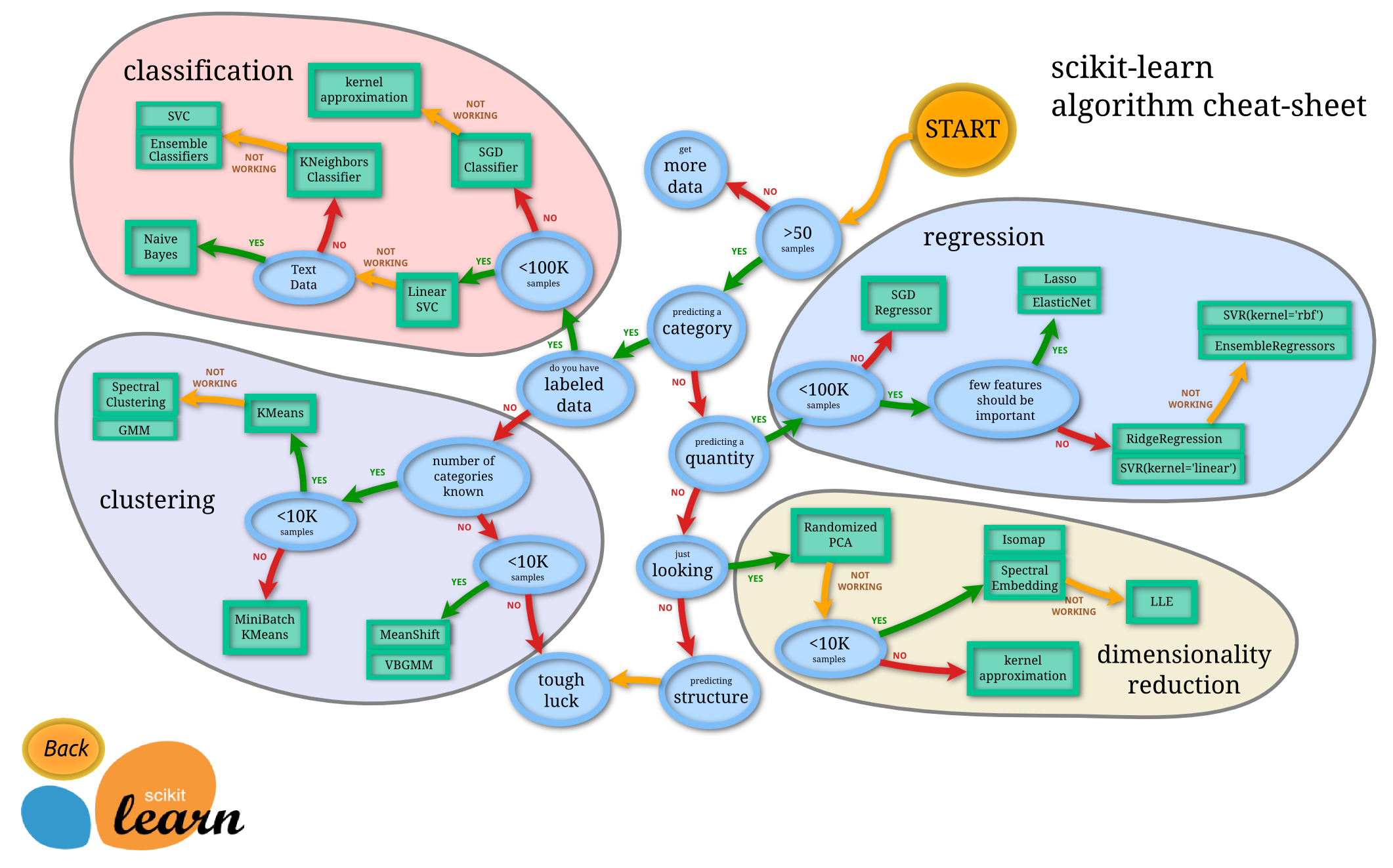
It's all about the API¶
sklearn is the defacto standard Machine Learning API for Python. Other libraries yield to the simplicity of its API.
- Want to do some Keras Deep learning? No problem, just use
keras.wrappers.scikit_learn - XGBoost anyone? Use:
xgboost.sklearn - Don't want to learn the syntax for the Light GBM?
lightgbm.sklearnto the rescue. - Natural langauge processing requires unique functionality, right? Nope,
nltk.classify.scikitlearn
Scikit-learn is a consistent API for all Machine Learning Algorithms¶
Estimators are the building block of scikit-learn. Almost everything is an estimator. All estimators have fit() methods. Most have either a predict() or transform() method. Supervised techniques generally have a score() method as well.
The basic ML workflow looks like this:
from sklearn.EstimatorFamily import Estimator
est = Estimator(hyperparameter_1, ... ,hyperparameter_n) # Create a model
est.fit(X_train, y_train) # Fit the model
est.score(X_test, y_test) # Evaluate model efficacy
est.predict(X_test) # Create predictions
Importing your estimators¶
from sklearn.EstimatorFamily import Estimator is typically how you'd import an estimator. Some examples are:
from sklearn.linear_model import RidgeRegression
from sklearn.ensemble import RandomForestRegressor, GradientBoostingClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.cluster import KMeans
from sklearn.neighbors import KNeighborsClassifier
from sklearn.neighbors import KNeighborsClassifier
Hyperparameters of your estimarors (Controlling how your estimator fits)¶
Instantiating an estimator typically looks like est = Estimator(hyperparameter_1, ... ,hyperparameter_n).
Upon instantition you have the option of setting hyperparameters (i.e. parameters whose values are set before the learning process). All hyperparameters have defaults that may or may not be satisfactory for your particular problem.
Exmaples of setting initial hyperparameters on an estimator:
rr = RidgeRegression(alpha=0.5, fit_intercept=False, normalize=True)
knc = KNeighborsClassifier(n_neighbors=10)
gbc = GradientBoostingClassifier()
Transformers - a special kind of estimator¶
Several sklearn estimators implement a transform() method. Transformers are typically used to 'transform' your featureset in a way that will improve another algorithms (e.g. regressor, classifier) performance.
Typical examples include:
sklearn.preprocessing.PCA # Principle Components transformation
sklearn.preprocessing.OneHotEncoder # Categorical to dummy transformation
sklearn.preprocessing.StandardScaler # Removing the mean and scaling to unit variance for each feature
sklearn.preprocessing.LabelEncoder # Single-column label to integer tranformation
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
response = ldf.index.get_level_values('LOB')
le.fit(response)
Mutating the Estimator with fit()¶
Though it looks like nothing happened, a lot happened under the hood. Our estimator has seen data can now be applied to new datasets. Once an estimator is fit, it spin off useful metadata that describes the fit model. sklearn uses a trailing underscore in property names to help users distinguish between hyperparameters and the new metadata.
from sklearn.linear_model import LinearRegression
lr = LinearRegression(fit_intercept=False)
lr.fit(X, y)
print(lr.fit_intercept) # A hyperparameter. Returns False.
print(lr.coef_) # Trailing underscore denotes the property comes from a 'fit'. Returns model coefficients.
Additionally the predict, transform, and score methods (if applicable) become available.
le.classes_
Supervised Learning Example - Identifying the line of business of an unlabeled triangle¶
We've computed the volume weighted development patterns of twenty companies for each line of business, wkcomp, comauto, and ppauto and want to use them to train a Machine Learning model that can identify the appropriate line of business.
Defining this problem more concretely:
The LDFs are our featureset, X, and the known line of business is our response, y.
X = ldf.values
y = le.transform(response)
Train/Test Split¶
It is best practice in machine learning to evaluate models on a test set of data. Since this is covered substantially in other literature, we will not go into the details of why here. sklearn comes with several utilities to split data, but we will explore the simplest one.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = \
train_test_split(X, y, test_size=0.33, random_state=42)
train_test_split returns a tuple of our features/response split into training and test sets. The random_state argument shows up in a lot of places in sklearn. Generally, when there is a stochastic component to the sklearn component you are using, random_state is there to allow you to set a seed so that your work can be replicated.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = \
train_test_split(X, y, test_size=0.33, random_state=42)
Fitting our classifier¶
Our data is in a numerical format, its been split, and now we are ready to do some Machine Learning.
Don't forget, when fitting any supervised learning technique, you must specify both your featureset and your response in the fit method.
model = KNeighborsClassifier()
model.fit(X_train, y_train)
model.score(X_test,y_test)
Trying another classifier¶
Remember the sklearn API was designed to make using different algorithms as consistent as possible. This means the same code should require minimal changes when applied to another classifier.
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from xgboost import XGBClassifier
from sklearn.linear_model import LogisticRegression, RidgeClassifier
for model in [RandomForestClassifier(n_estimators=10),
GradientBoostingClassifier(),
XGBClassifier(),
LogisticRegression(solver='lbfgs', multi_class='auto'),
RidgeClassifier(),
KNeighborsClassifier()]:
model.fit(X_train, y_train)
print(f'{model.__class__.__name__} holdout accuracy:',
model.score(X_test,y_test))
Cross-validation¶
sklearnprovides across_val_scoreto test the accuracy of an estimator across multiple folds painting a truer picture of an estimators' efficacy than a simple train/test split.- With
cross_val_score, we don't really need to provide separate train and test sets. Though, with enough data, it is sometimes instructive to have train/test and holdout
from sklearn.model_selection import cross_val_score
import numpy as np
knn = KNeighborsClassifier()
np.mean(cross_val_score(knn, X, y, cv=5))
Improving model accuracy with GridSearchCV¶
With GridSearchCV, we can feed a hyperparameter grid into our estimator to determine an 'optimal' set of hyperparameters to use for our particular business problem. GridSearchCV itself is an estimator and so it has the usual 'fit() and predict() methods any other classifier would.
At a minimum, parameterizing the GridSearchCV estimator we need to specify:
- The estimator we want to use
- The hyperparameter searchspace as a dictionary
Optionally, we can also specify:
- The number of folds to use
from sklearn.model_selection import GridSearchCV
param_grid=dict(n_neighbors=[1,3,5,7,9,11], p=[1,2,3,4,5,6])
grid = GridSearchCV(knn, param_grid, cv=5)
grid.fit(X, y)
print(f'Best Score: {grid.best_score_}')
grid.best_estimator_
Holding p=3 constant, a visual inspection of the cross-validated scores shows support for n_neighbors=3
import seaborn as sns
sns.set_style('whitegrid')
p_3 = grid.cv_results_['param_p']==3
g = sns.pointplot(x=grid.cv_results_['param_n_neighbors'][p_3],
y=grid.cv_results_['mean_test_score'][p_3]) \
.set(xlabel='n_neighbors', ylabel='Accuracy', title='Gridsearch Results')
Confusion Matrix¶
It looks like our classifier struggles more to distinguish between private passenger auto and workers' compensation.
from sklearn.metrics import confusion_matrix
pd.DataFrame(confusion_matrix(y, grid.best_estimator_.predict(X)),
index=le.classes_, columns=le.classes_)
Visual representation of the data¶
By inspection (at least across the first three development ages), it is more difficult to distinguish between wkcomp and ppauto in line with where our classifiers are least accurate.
plot_data=ldf.reset_index().iloc[:,1:].set_index('LOB').T
g = sns.pairplot(ldf.reset_index()[['LOB','1-2','2-3','3-4']], hue="LOB")
More complex workflows with Pipeline¶
The authors of sklearn recognize that composability of multiple estimators will be necessary to build the best models. For example, you may want to cluster a feature before feeding it into a Regressor.
The Pipeline is useful for chaining one or more transformers together. Pipelines themselves are estimators and have fit(), predict(), and score() function and can be used with all of the sklearn funcitons used for regular estimators including but not limited to: cross_val_score, confusion_martix
Adding a PCA step with no hyper-parameter tuning actually reduces our cross validation accuracy score.
from sklearn.pipeline import Pipeline
from sklearn.decomposition import PCA
steps=[('pca', PCA()),
('knn',KNeighborsClassifier(n_neighbors=3, p=3))]
pipe = Pipeline(steps=steps)
np.mean(cross_val_score(pipe, X, y,cv=5))
Can we do better with parameter tuning?¶
Pipelines and GridSearchCV¶
Since a Pipeline is just another estimator GridSearchCV allows the hyperparameter space of all estimators in the pipeline to be gridsearched in one go.
To avoid hyperparameter name clashes between one estimator and another within a pipeline, sklearn uses a double underscore naming convention of the form {estimator_name}__{hyperparameter} for the keys of its parameter grid.
We achieve parity with the highest accuracy of our original classifier. In this instance, adding the Principle Components step did not yield any better results.
param_grid=dict(knn__n_neighbors=[1,3,5,7,9,11],
knn__p=[1,2,3,4,5,6],
pca__n_components=[3, 5, 7, 9])
pipe = Pipeline(steps=[('pca', PCA()),
('knn',KNeighborsClassifier())])
grid = GridSearchCV(pipe, param_grid, cv=5, refit=True)
grid.fit(X, y)
print(f'Best Score: {grid.best_score_}')
grid.best_estimator_
Scikit-Learn Recap¶
- Almost everything is an Estimator. They all have a
fitmethod and depending on the nature of the estimator may also have apredict,scoreortransformmethod. - The API is standardized across estimator
- A transformer is a special type of estimator that trasnforms data for another Estimator
- Cross-validation with Grid Search helps in hyperparameter selection
- Pipelines are useful for composing a chain of Estimators.
- The documentation is a goldmine of information